Once an alignment has been partitioned (see working with partitions for further information), TOPALi can use this information in an attempt to determine which sequences within the alignment are recombinants (mosaics) and how they relate to the pure (non mosaic) strains..
TOPALi does this by creating partition sequences, which are special sequences formed by taking each mosaic sequence and splitting it up into a several new sequences - one sequence per partition. For example, if the following sequence was known to be mosaic:
SeqA AACGTGCATGCG
and three partitions had been found (with breakpoints at 33% and 66% of the sequence length), then the following three partition-sequences would be created from it:
SeqA_P1 AACG????????
SeqA_P2 ????TGCA????
SeqA_P3 ????????TGCG
TOPALi then fits these sequences and the original non-mosaic sequences onto a tree, whose distance matrix can be used to determine sequence similarity.
Start the wizard by selecting Analysis | Predict Mosaic Structure from the menu bar. This brings up the Predict Mosaic Structure Wizard dialog. The first stage of the process is to determine which sequences are the mosaics.

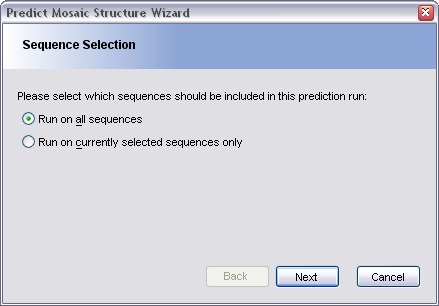
In the Sequence Selection pane, choose whether to run the mosaic structure prediction algorithms on all sequences in the alignment, or on just the currently selected sequences.
In the Additional Options pane, set the divergence threshold to use (which determines how close to a pure strain a mosaic partition must be for it to be considered similar to it), and also choose whether to add the newly created partition-sequences into the original alignment, and whether to include the mosaic sequences (in addition to the partition-sequences derived from them) in the final tree calculations.
In the Determine Mosaic Sequences pane, choose whether to have TOPALi automatically estimate which sequences are mosaic, or select to choose them manually. Automatically predicting which sequences are mosaic may take some time. Regardless of the method chosen, once complete, the final pane of the dialog is displayed:
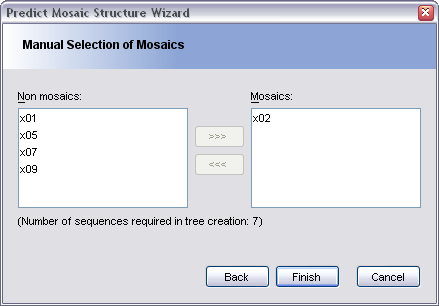
This pane allows you to either confirm the selection of mosaics made by TOPALi, or to select them yourself. Sequences marked as mosaic should appear in the right-hand box, whereas sequences marked as non-mosaic should remain in the left-hand box.
Click Finish to confirm the settings, and TOPALi will create the reconstituted tree that is used to perform the final similarity analysis.
Once TOPALi has made a mosaic structure prediction, the results will appear in a new Mosaic Structure tab.
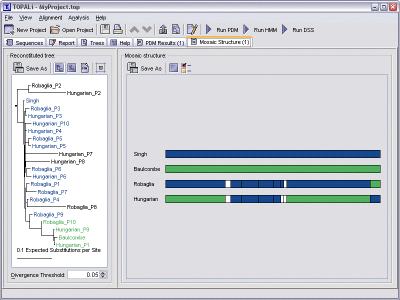
This view is split into two sections.
The left-hand side displays a reconstituted tree, formed during the mosaic structure prediction using the partitions and mosaic sequence information that was supplied (either determined by TOPALi or selected by the user). This tree can be interacted with in the same way as all trees in TOPALi - see viewing trees for further information on this.
The right-hand side displays the actual mosaic structure. Each sequence in the alignment is displayed as a coloured bar - non mosaics are formed from a single solid colour, whereas mosaics (sequences containing recombinant events) will be displayed using the appropriate colours for the partition in question.
Hovering the mouse over any sequence will display more information on the partition under the mouse - in the status bar and as a tool tip.
The mosaic structure diagram can be saved in one of three formats:
To save the diagram, click the Save Mosaic Structure As button.
|
|
Save Mosaic Structure As |
The Save Mosaic Structure As dialog will then appear, allowing you to choose a location on disk for the structure, along with the data format to save it in.
The bars displayed within the structure can have simple gradient shading applied to them. To toggle this on and off, press the Set Gradient Shading toolbar button.
|
|
Set Gradient Shading |
TOPALi randomly selects the colours to be used for each unique sequence (based on the settings specified in the Display Settings dialog. See display settings for further information).
However, you can override these colours by manually clicking on any sequence's colour bar. This will bring up a colour chooser allowing you to select a new colour to display that sequence with. Any mosaic sequences using the same colour will also update to use the new colour (as will the appropriate non-mosaic sequence if the original sequence clicked upon was a mosaic).