TOPALi provides the following (minimal) set of alignment editing features.
The display order of sequences can be reordered to aid visual navigation of an alignment file. To do this, first select the sequences you want to move by left-clicking the mouse on their names within the Sequences pane (more information on this pane is available here). Hold down CTRL if you want mutliple sequences selected while doing this.
The sequences can then be moved up or down the display area by selecting Alignment | Move Sequences Up or Alignment | Move Sequences Down from the menu bar, clicking the Move Selected Sequences Up (or Down) toolbar buttons, or by pressing Alt-Up or Alt-Down on the keyboard.
|
|
Move Selected Sequences Up |
|
|
Move Selected Sequences Down |
You can also move a group of sequences to the top of an alignment by selecting Alignment | Move Sequences to Top from the menu bar.
Note than in addition to visually reordering the sequences, their new positions within the alignment will be reflected in any output files that are exported from the project, such as saving the alignment under a new name or in a different alignment format.
To rename a sequence, first select it from the sequence list with the mouse, then select Alignment | Rename Sequence from the menu bar. Note that TOPALi will only accept sequence names that do not contain space characters.
To replace characters within an alignment, select Alignment | Edit Alignment | Replace Characters from the menu bar. This brings up the Replace Characters dialog.
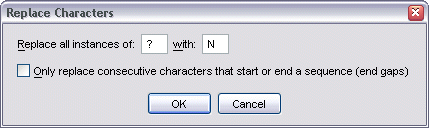
By default, this dialog allows you to replace all instances of a character within the alignment with another character. However, by ticking the Only replace... option, TOPALi will only replace characters that form part of a consecutive sequence that either start or end a sequence.
As an example, consider the following simple alignment data set:
AACCTT??CGTC
???CTT??GAT?
If the Only replace... option is not ticked, then all instances of ? would be replaced with N, resulting in:
AACCTTNNCGTC
NNNCTTNNGATN
However, if the option is ticked, then the data set becomes:
AACCTT??CGTC
NNNCTT??GATN
To remove gap characters within an alignment, select Alignment | Edit Alignment | Remove Gaps from the menu bar. This brings up the Remove Gaps dialog.
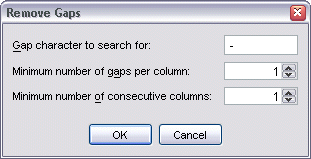
The minimum number settings allow you to specify in greater detail which gaps should be removed.
Note that once gaps have been removed from an alignment, all PDM, HMM, and DSS analysis result graphs that were generated from the alignment before the gaps were removed will be disabled until the gaps are restored. This is because the graphs (and any reports generated using them) depend on data from the original alignment that may no longer be present.
Note also that gaps can only be removed once, even though the parameters specified in the dialog may not remove all gap characters. To remove further gaps, those that were removed must first be restored (see below), then the Remove Gaps dialog re-run with new parameter settings.
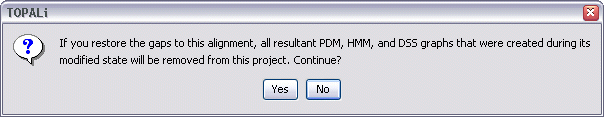
To restore previously removed gaps, select Alignment | Edit Alignment | Restore Gaps from the menu bar.
Note that this operation is non-reversible, that is, all PDM, HMM, and DSS analysis results graphs that were generated while gaps were removed from the alignment will be lost if those gaps are restored.
Partition-sequences (sequences that were created as part of the process of determining the mosaic structure of the alignment), can be removed by selecting Alignment | Edit Alignment | Remove Partition-Sequences from the menu bar.
See predicting mosaic structure for more information.