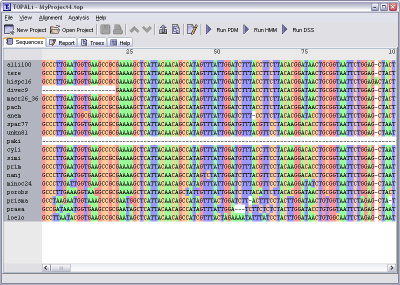
TOPALi displays alignments within its Sequences pane.

The main section of this view provides information on the actual sequence data (colour coded to the settings specified within the Display Settings dialog). Moving the mouse over any site will provide its exact column number in the status bar along the bottom, whereas hovering the mouse over a site will provide this information as a tooltip.
Further information to help with viewing the data is provided along the top of the sequence area, showing both site positions (every 25 base pairs), as well as ClustalX-style '*' information that prints a '*' whenever every nucleotide in a column is identical.
Each sequence's name is displayed down the left-hand side of the pane. When a new project is created, all sequences will be selected by default, but sub-selections can also be picked manually with the mouse.
Scroll bars are available whenever the alignment is too large to fit within the viewing area, allowing the view to be moved left and right, or up and down, in order to view other areas of the alignment.